CellularPotts. jl: simulating multiscale cellular models in Julia
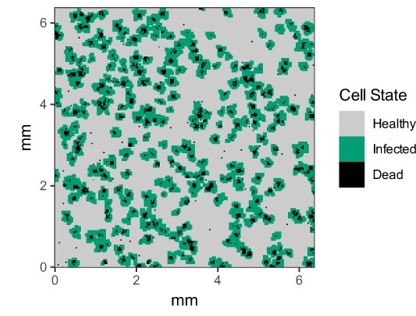
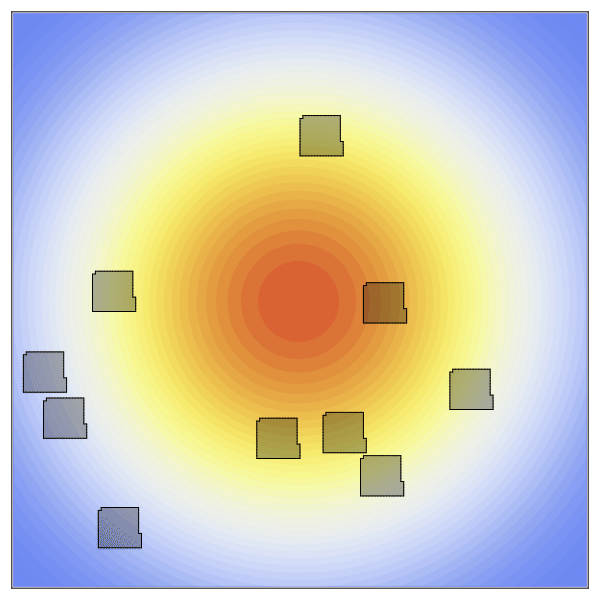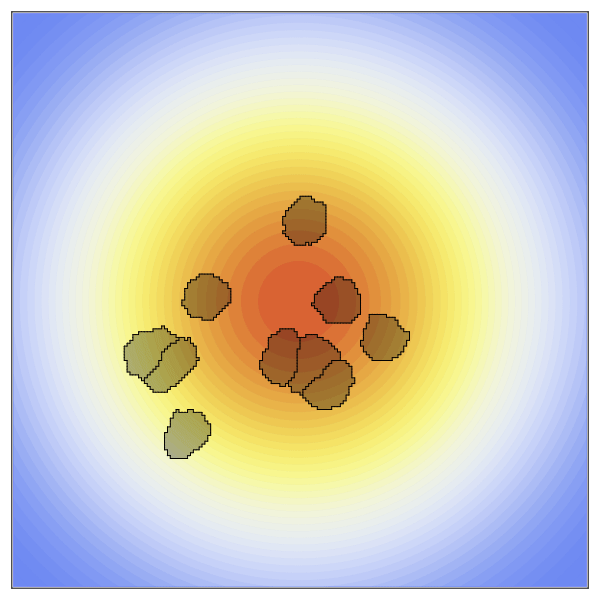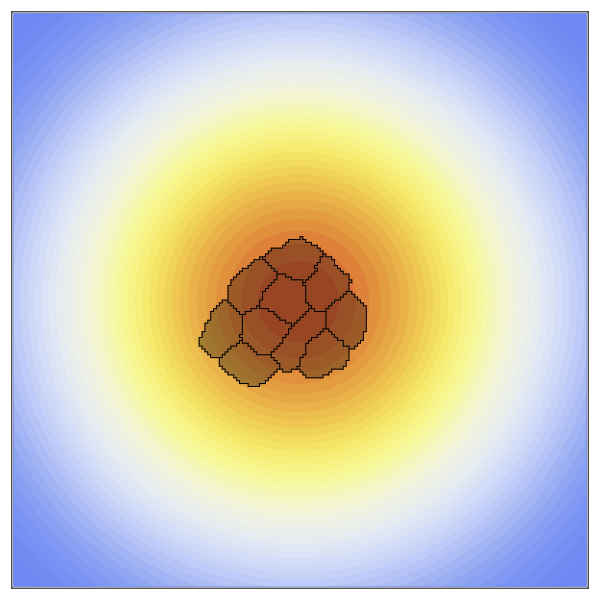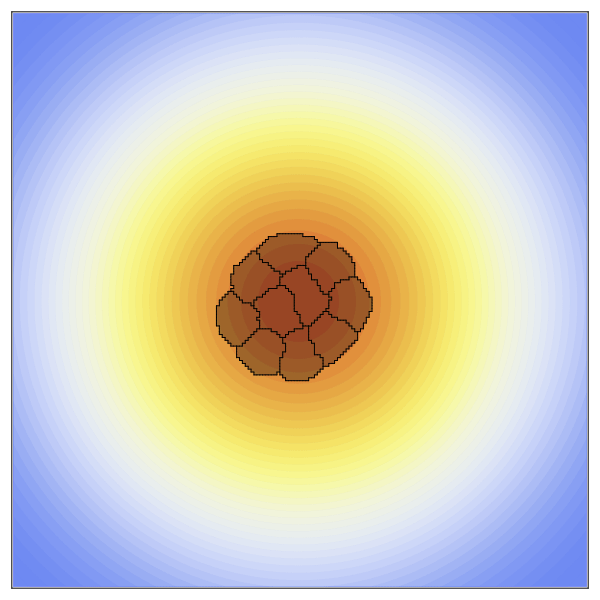
CellularPotts.jl is a software package written in Julia to simulate biological cellular processes such as division, adhesion, and signaling. Accurately modeling and predicting these simple processes is crucial because they facilitate more complex biological phenomena related to important disease states like tumor growth, wound healing, and infection. Here we take advantage of Cellular Potts Modeling to simulate cellular interactions and combine them with differential equations to model dynamic cell signaling patterns. These models are advantageous over other approaches because they retain spatial information about each cell while remaining computationally efficient at larger scales. Users of this package define three key inputs to create valid model definitions: a 2- or 3-dimensional space, a table describing the cells to be positioned in that space, and a list of model penalties that dictate cell behaviors. Models can then be evolved over time to collect statistics, simulated repeatedly to investigate how changing a specific property impacts cellular behavior, and visualized using any of the available plotting libraries in Julia.
DOI Link