On Patrol
Here we combine ideas from the HelloWorld.jl and LetsGetMoving.jl examples to simulate T-Cells patrolling through a dense layer of epithelial cells.
Start by loading in the CellularPotts.jl package and creating a space where cells can exist.
using CellularPotts
space = CellSpace(200,200; diagonal=true)200×200 Periodic 8-Neighbor CellSpace{Int64,2}Initialize a new CellState with 75 epithelial cells and 5 T-Cells
initialCellState = CellState(
names = [:Epithelial, :TCell],
volumes = [250, 200],
counts = [160, 10]);Note that for the MigrationPenalty we set the epithelial cell's scaling factor to zero. This effectively removes this penalty from the cell type.
penalties = [
AdhesionPenalty([30 30 30;
30 20 30
30 30 40]),
VolumePenalty([30, 30]),
PerimeterPenalty([0, 5]),
MigrationPenalty(75, [0, 100], size(space))
]4-element Vector{Penalty}:
Adhesion
Volume
Perimeter
MigrationCreate a new CellPotts model.
cpm = CellPotts(space, initialCellState, penalties)Cell Potts Model:
Grid: 200×200
Cell Counts: [Epithelial → 160] [TCell → 10] [Total → 170]
Model Penalties: Adhesion Volume Perimeter Migration
Temperature: 20.0
Steps: 0Finally, lets run the model for a few steps to let the initial cell positions to equalibrate.
for i=1:50
ModelStep!(cpm)
endRecord the simulation. Here we also show how to customize the cell colors, run the recording for more time steps, and skip frames.
record(cpm; file="OnPatrol.gif", cellcolors=[:grey90, :lightblue], steps=1500, skip=10)
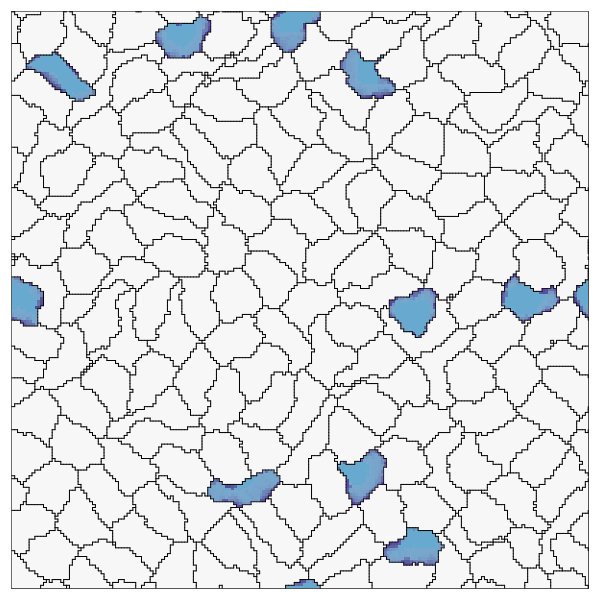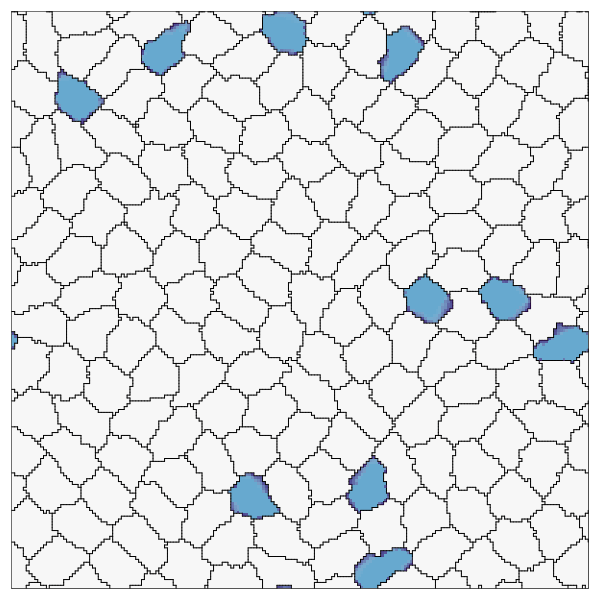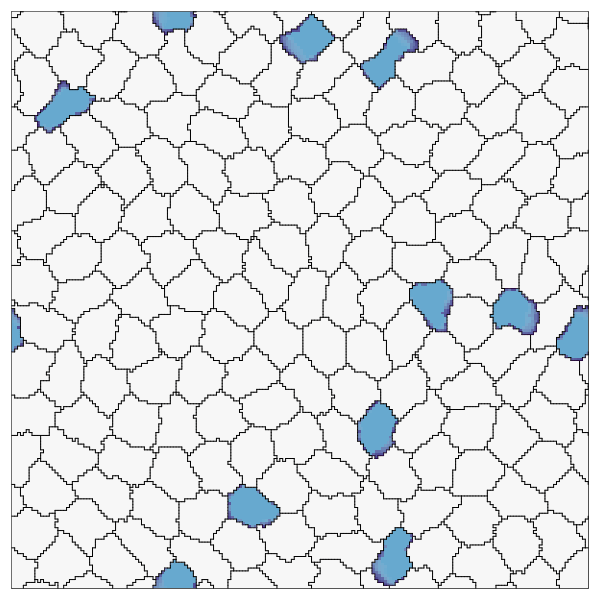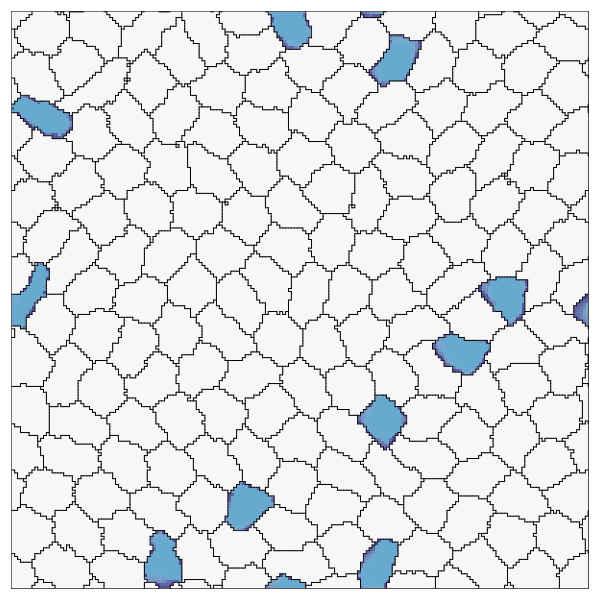
This page was generated using Literate.jl.