Over Here!
In this example we'll demonstrate how to create a static gradient field that attracts cells to its center.
We'll start by importing packages and specifying dimensions for the space cells will occupy. Here those dimensions are declared constants because they will be used repeatedly.
using CellularPotts, Plots
const xdim = 200
const ydim = 200
space = CellSpace(xdim, ydim, periodic=false, diagonal=true)200×200 nonPeriodic 8-Neighbor CellSpace{Int64,2}Initialize 10 cells randomly positioned.
initialCellState = CellState(names=:TCells, volumes=200, counts=10)┌─────┬────────┬─────────┬─────────┬─────────┬────────────────┬────────────┬───────────────────┐
│ Row │ names │ cellIDs │ typeIDs │ volumes │ desiredVolumes │ perimeters │ desiredPerimeters │
│ │ Symbol │ Int64 │ Int64 │ Int64 │ Int64 │ Int64 │ Int64 │
├─────┼────────┼─────────┼─────────┼─────────┼────────────────┼────────────┼───────────────────┤
│ 0 │ Medium │ 0 │ 0 │ 0 │ 0 │ 0 │ 0 │
│ 1 │ TCells │ 1 │ 1 │ 0 │ 200 │ 0 │ 168 │
│ 2 │ TCells │ 2 │ 1 │ 0 │ 200 │ 0 │ 168 │
│ 3 │ TCells │ 3 │ 1 │ 0 │ 200 │ 0 │ 168 │
│ 4 │ TCells │ 4 │ 1 │ 0 │ 200 │ 0 │ 168 │
│ 5 │ TCells │ 5 │ 1 │ 0 │ 200 │ 0 │ 168 │
│ 6 │ TCells │ 6 │ 1 │ 0 │ 200 │ 0 │ 168 │
│ 7 │ TCells │ 7 │ 1 │ 0 │ 200 │ 0 │ 168 │
│ 8 │ TCells │ 8 │ 1 │ 0 │ 200 │ 0 │ 168 │
│ 9 │ TCells │ 9 │ 1 │ 0 │ 200 │ 0 │ 168 │
│ 10 │ TCells │ 10 │ 1 │ 0 │ 200 │ 0 │ 168 │
└─────┴────────┴─────────┴─────────┴─────────┴────────────────┴────────────┴───────────────────┘
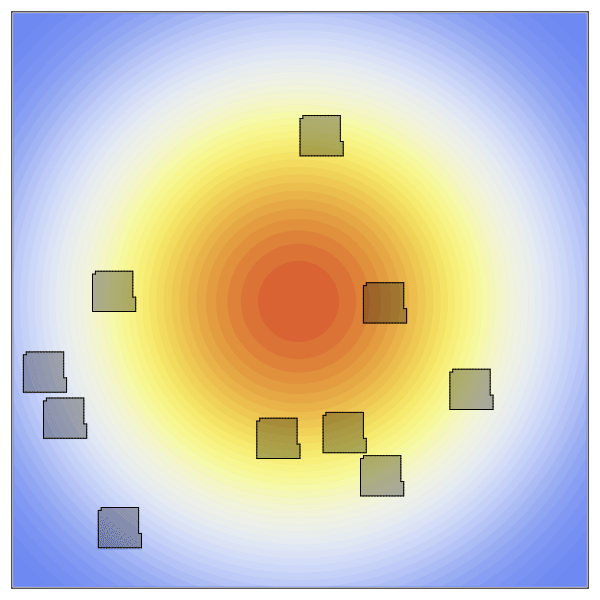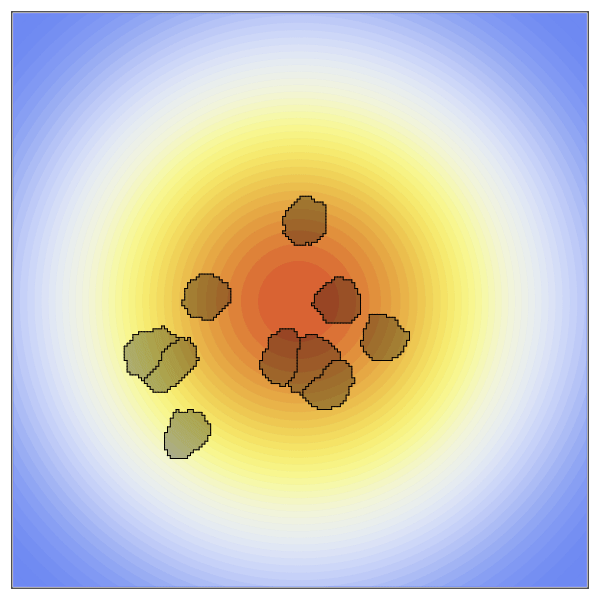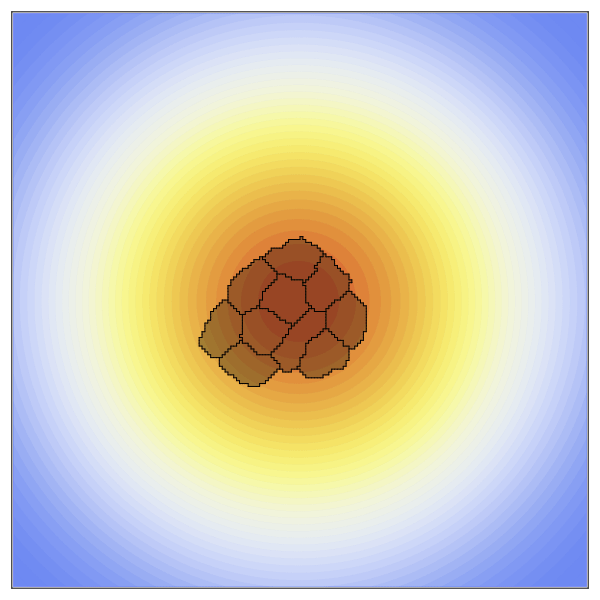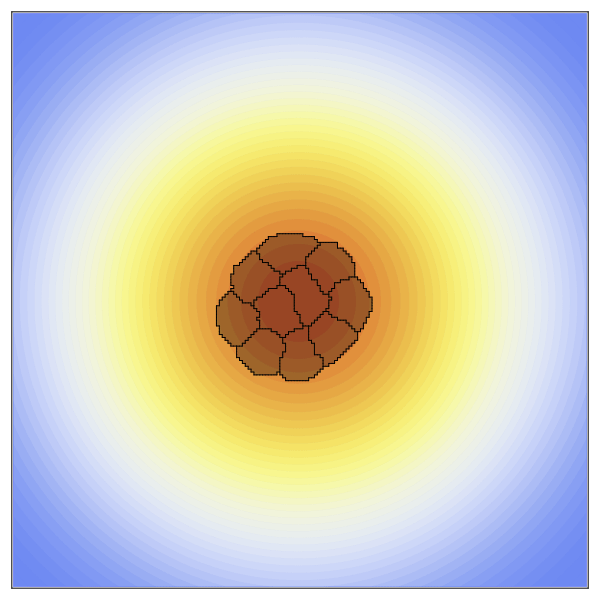
Now to create the static concentration gradient. Higher values will be positioned in the center of the space and expoentially drop off as it moves towards the edges.
species = [100exp(-((x-xdim/2)^2+(y-ydim/2)^2)/10000) for x in 1:xdim, y in 1:ydim];The new penalty added here is the ChemoTaxisPenalty. This penalty takes two arguments, the strength of the chemoattraction and the concentration gradient. Here we create a strong gradient by providing a large scaling factor.
penalties = [
AdhesionPenalty([30 30; 30 30]),
VolumePenalty([10]),
ChemoTaxisPenalty([50], species)
];Now we can take these three objects and create a Cellular Potts Model object.
cpm = CellPotts(space, initialCellState, penalties)
anim = @animate for t in 1:2000
plt = contourf(species,
c=:temperaturemap,
levels=50,
alpha=0.2,
linewidth=0,
legend=false)
ModelStep!(cpm)
visualize!(plt, cpm; cellcolors = RGBA(0,0,0,0.3))
end every 10
gif(anim, "OverHere.gif", fps = 60)
This page was generated using Literate.jl.